Abstract
Introduction:
Diffuse large B-cell lymphoma (DLBCL) is the most commonly occurring lymphoid malignancy in the United States. Through gene expression profiling (GEP), two biologically distinct molecular subgroups of DLBCL have been identified - germinal center B-cell (GCB) and activated B-cell - with disparate prognostic implications. Because GEP is not yet easily implemented in clinical practice, immunohistochemical (IHC) algorithms have been developed and validated to segment DLBCL into cell-of-origin subtypes. The most commonly employed algorithm uses CD10, BCL6 and MUM1 to distinguish GCB and non-GCB subtypes, but has been shown to misclassify up to 20% of cases. Advancements in digital pathology technology have led to the development of image analysis algorithms that allow for the extraction of quantitative descriptions of pathology image features such as level of IHC staining. We explore the feasibility of developing a precise, repeatable, and objective image analysis-based scoring algorithm for DLBCL whole slide images to replicate DLBCL subtype classification and prognostic assessment.
Methods:
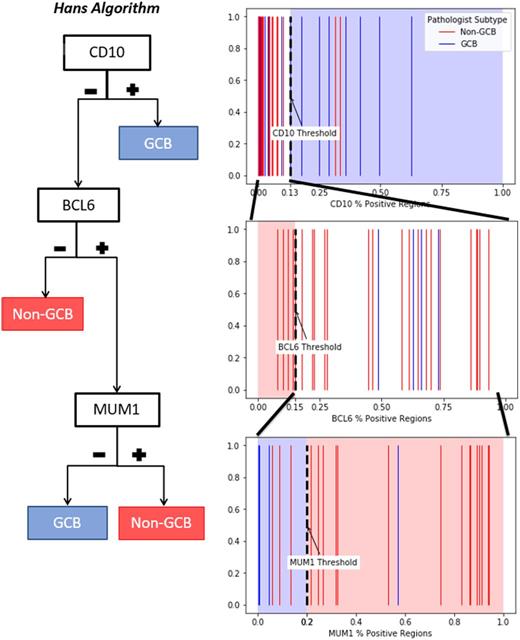
Tissue slides of the immunostains included in the Hans algorithm (CD10, BCL6, MUM1) for 40 DLBCL patients were digitized at 40x objective resolution using Hamamatsu Nanozoomer 2.0 HT or Aperio AT2. We developed and trained an image analysis algorithm on the IHC whole slide images. The algorithm first masks the tissue area of the whole slide image, then performs superpixel segmentation, dividing tissue area into compact regions (60 microns square). Each region is normalized to a reference IHC image and has a color deconvolution algorithm applied to separate the hematoxylin and diaminobenzidine (DAB) stains. Features describing the distribution of both the hematoxylin and DAB stains are extracted using convolutional autoencoders. We then trained a classification algorithm using these features to classify the staining of these regions as positive or negative using machine learning techniques. Whereas pathologists examine specific regions in a high-power field and use visual estimation to determine if >30% of tumor cells are positive, our algorithm measures the percentage of positive regions over the entire DLBCL tissue. Receiver operating characteristics (ROC) curves were calculated to assess ability of image analysis-based measurements to predict pathologist classifications as positive or negative for CD10, BCL6, and MUM1. Pathologist classification of DLBCL subtype and positive or negative for each immunostain were obtained from pathology reports. Using thresholds for percent positive regions established by the ROC curves, patients were classified into GCB/non-GCB by the Hans algorithm using sequential application of the identified thresholds as shown in the Figure. The regions in the figure define the algorithm classification and the lines correspond to the pathologist classification displaying GCB in blue and non-GCB in red. Concordance with pathologist subtyping was calculated by agreement rate and Cohen's κ statistic.
Results:
Of the 40 patients included in our study, pathologists classified CD10, BCL6, and MUM1 as positive in 27.5%, 90%, and 84.9% respectively. Area under the ROC curve (AUC) for predicting pathologist classifications of positivity from image analysis measurements for CD10, BCL6, and MUM1 were 0.92, 1.0, and 0.95 respectively. Thresholds from the ROC curves for CD10, BCL6, and MUM1 were 13%, 15%, and 20% respectively. Using these thresholds, classification by image analysis algorithm was concordant with pathologist classification in 82.5% (κ = 0.65) of cases.
Discussion:
Pathologist IHC classification is limited by interobserver and interlaboratory variability, human error, difficulties with human eye distinguishing between low-intensity stains, and ordinal outputs. These limitations have led to inconsistent prognostic significance with IHC cell-of-origin subtyping. We developed a novel, objective image analysis scoring for predicting DLBCL subtype that uses whole slide images. Our image analysis algorithm may provide an effective decision-support tool for pathologist subtype classification and address known limitations. Future work will compare subtyping by the image analysis algorithm to gene expression profiling and inter-rater reliability of tissue microarrays and whole slide images performed by expert hematopathologists.
Flowers: National Institutes Of Health: Research Funding; Burroughs Welcome Fund: Research Funding; Celgene: Consultancy, Research Funding; Gilead: Consultancy; Research to Practice: Research Funding; Infinity: Research Funding; Onyx: Research Funding; Seattle Genetics: Consultancy; V Foundation: Research Funding; Genentech/Roche: Consultancy, Research Funding; OptumRx: Consultancy; Bayer: Consultancy; Millennium/Takeda: Research Funding; Eastern Cooperative Oncology Group: Research Funding; Clinical Care Options: Research Funding; National Cancer Institute: Research Funding; TG Therapeutics: Research Funding; Acerta: Research Funding; Prime Oncology: Research Funding; Educational Concepts: Research Funding; Spectrum: Consultancy; Pharmacyclics LLC, an AbbVie Company: Research Funding; Janssen Pharmaceutical: Research Funding; Abbvie: Consultancy, Research Funding. Cooper: Varian Medical Systems: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal